In the same way that its older brother SUIT, SUIT-N toolbox was written to support a number of different analysis techniques, such as volumetry and fMRI group analysis. All techniques share a number of initial similar steps, including the isolation of the cerebellum from the rest of the brain, normalization to the atlas template, and reslicing of the data into atlas space.
Isolate (suit_isolate_neo)
The Isolation algorithm works best on a whole-brain high-resolution (~1 mm isotropic) T2 scan with good gray-white matter contrast. The algorithm works best if the image is brought into LPI- or RPI-orientation, and the origin of the image is set to the anterior commissure.
To isolate the cerebellum simply type suit_isolate_neo({'<name>.nii'}) at the matlab
command window. The isolation procedure:
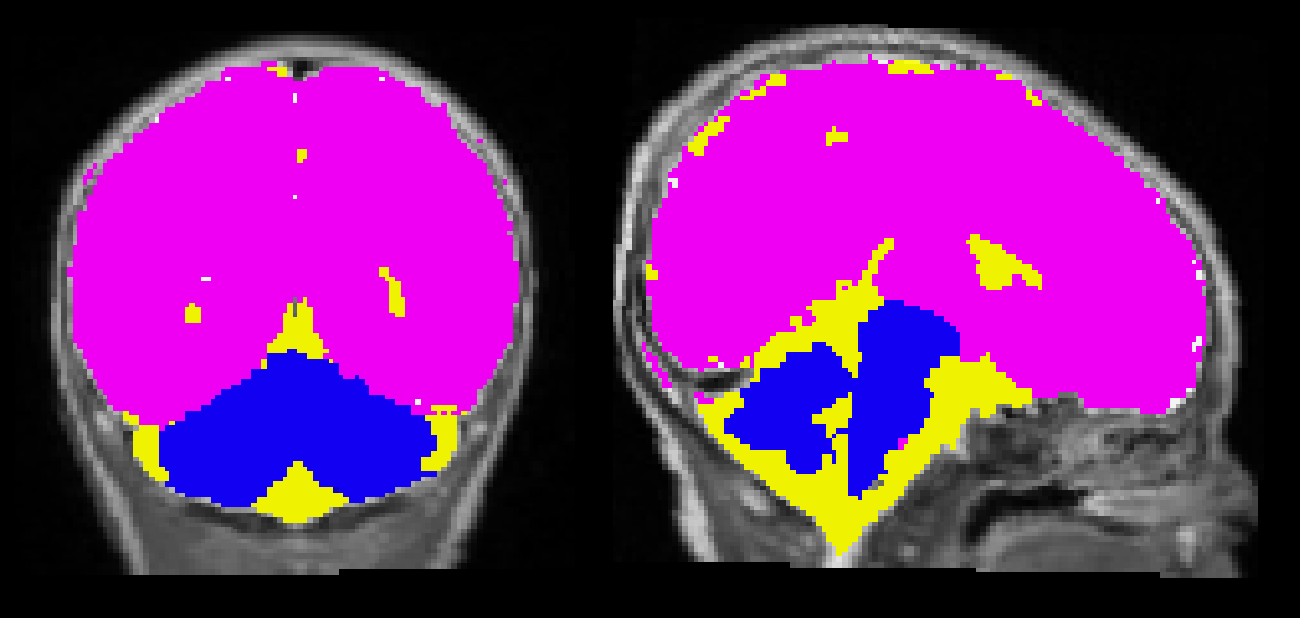
- segments the brain into tissue-types using John Ashburner's segmentation algorithm
- crops the full volume, such that it mostly includes the
infra-tentorial structures and saves the cropped image as
c_<name>.img - uses the tissue-type and the proximity to cortical white matter to calculate the posterior probability of each voxel to belong to cerebellum or brainstem.
- This cerebellar mask is saved as
c_<name>_pcereb.nii. - You should check the isolation map visually and if necessary, hand- correct it using a image viewer (e.g. MRICroN, CARET, FSL view)
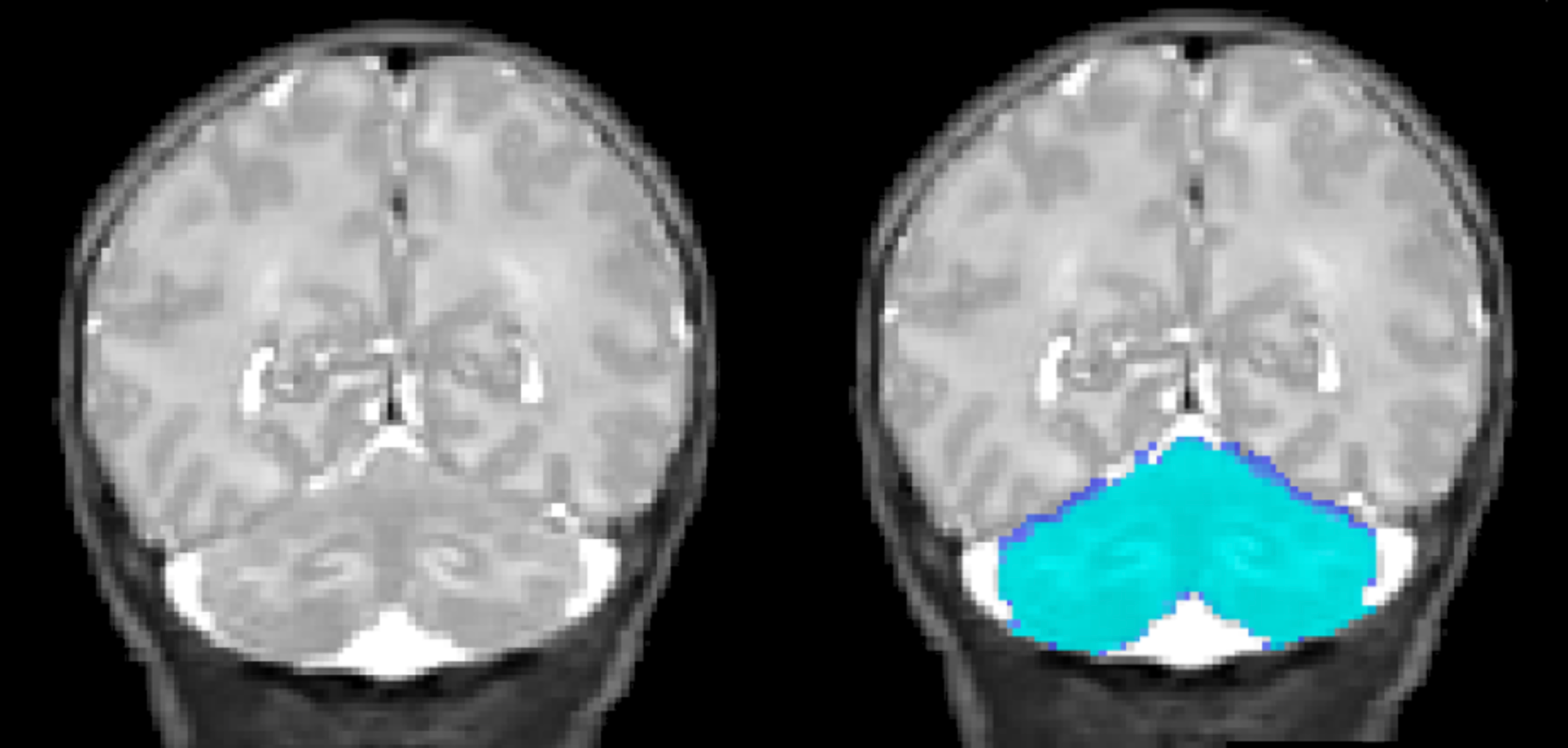
Cerebellar isolation map generated by the isolation algorithm
Cerebellar segmentation generated by the isolation algorithm
Isolation takes about 2 min on a normal desktop PC. Even if
hand correction is necessary, it does not take much more than
5-10 min per individual. Additionally, the function can return the volume of the cerebellum
and ICV using the "getV" flag. For full options of the isolation algorithms
from the command line type: help suit_isolate_neo.
Normalize (suit_normalize_neo)
To normalize a cerebellum to the SUIT-N atlas template typesuit_normalize_neo in the matlab command
window, you should then select the cropped image
(c_<name>.nii) and the (possibly handcorrected) isolation map
(c_<name>_pcereb_corr.nii)
as a mask using the SPM interface. Both of these files are generated
in the cerebellum isolation stage listed directly above.
- The normalization procedure finds a nonlinear deformation map to the SUIT template, using the cosine-basis function approach (Ashburner & Friston, 1999).
- The deformation is saved as a mat-file under
mc_<name>_snc.mat. - A resliced version of the masked, normalized anatomical image is saved under
wsuit_mc_<name>_snc.mat.
For full options of the normalization process type: help suit_normalize_neo.
Reslice images into SUIT space (suit_reslice_neo)
The function suit_reslice_neo uses the deformation map found
in the normalization step to resample images into the new atlas
space. To reslice an image into atlas space simply type suit_reslice_neo in the matlab command
window including the appropiate image to be resliced (or a cell array if several), the deformation map
mc_<name>_snc.mat and the isolation map c_<name>_pcereb_corr.nii.
A full call to the function would therefore look like:
suit_reslice_neo({'<name>.nii'},c_<name>_pcereb_corr.nii,mc_<name>_snc.mat). For
full options of the function see: help suit_reslice_neo.
Reslice images into subject space (suit_reslice_inv)
The function suit_reslice_inv takes an image in SUIT or SUIT-N space and
reslices it into the space of the individual subject.
This can be especially useful when a region of interest or a set of areas are defined in the SUIT-N
space to be resliced after in to the original space.
To reslice an image into atlas space simply type suit_reslice_inv. Then select the
appropriate image in SUIT-N space to as
well as the deformation map ( mc_<name>_snc.mat).
For full options of the function see: help suit_reslice_inv.